ETL Pipeline
ETL stands for Extraction, Transformation, Loading. We have an ETL pipeline for taking in the RAW videos given to us by the hospital, extracting and storing the frames from it, and maintaining CSV files pointing to these frames with the frame status, such as timestamp, disturbed, etc. This allows us to take the information from the CSVs and make a dataset using them that contains paths to the previously extracted frames instead of re-extracting the frames again from the videos, which is computationally very expensive. All the ETL pipeline, data extraction and modification happens in the ‘trishul’ server. The ETL Pipeline contains these parts:
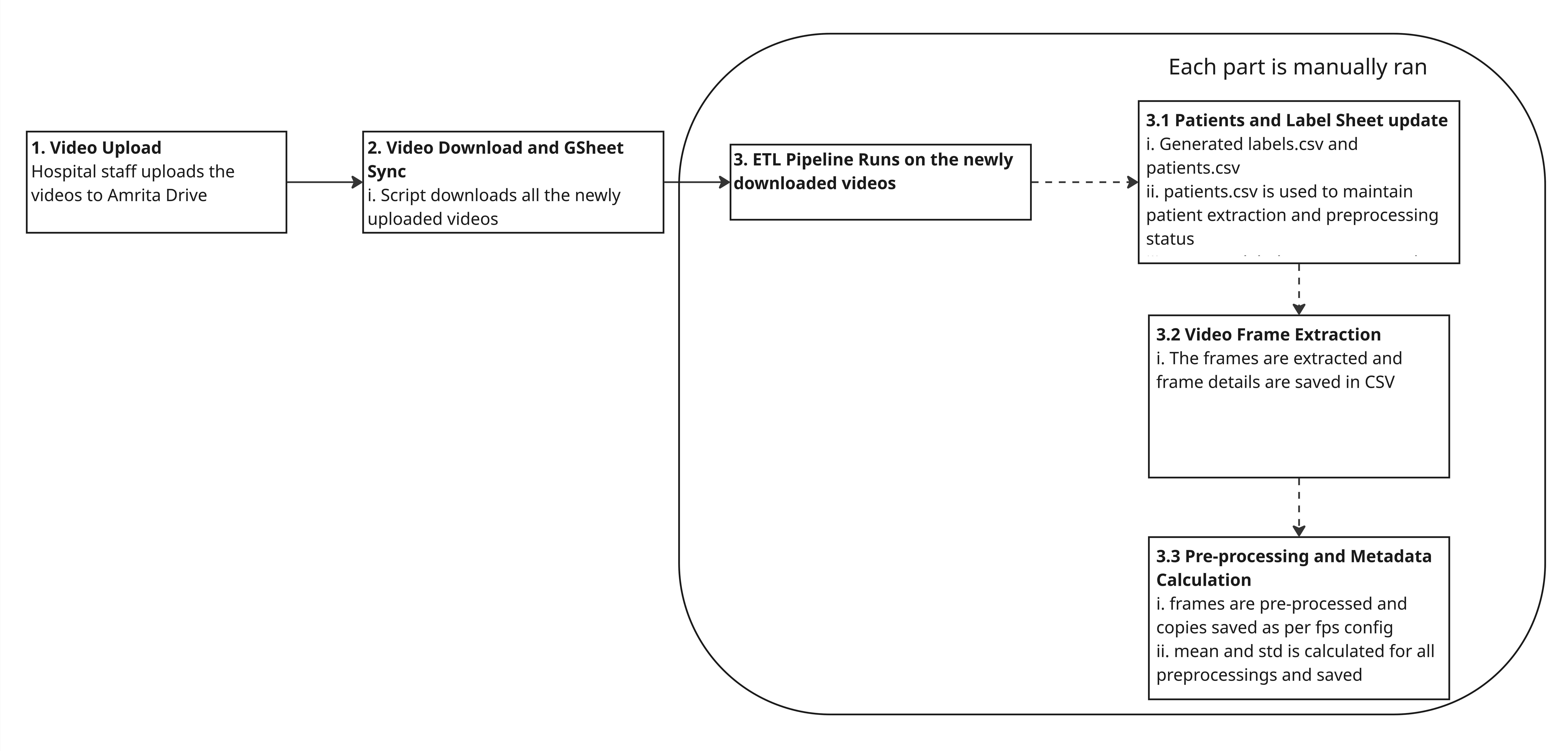
- Video Upload: Hospital Staff uploads the videos to Amrita Drive
-
Video Download and Gsheet Sync: Script downloads all newly uploaded videos and runs at 5am every morning. The script is stored at https://github.com/Amrita-Medical-AI/server-infrastructure/blob/main/eusml-fatty_liver-drive-sync.sh and refer to the documentation of the https://github.com/Amrita-Medical-AI/server-infrastructure repo to get it working.
The downloaded videos at the time of writing is saved at
/data/eusml/raw_datafor EUSML and at/data/fatty_liver/raw_data. The other configs can be found in theeusml-fatty_liver-drive-sync.shscript. 3. ETL This is the mian part of the ETL Pipeline. Code is stored at https://github.com/Amrita-Medical-AI/eusml-ETL-pipeline3.1 Patients and Label sheet Update We stored the time stamp labels from DynamoDB after processing it in labels.csv. The patient specific data like cancer type, cap score etc are stored in patients.csv. These files are present in
/data/{eusml, fatty_liver}/master-dataset/Data.Run
labels_generator/{eus,liver}_generator.py_3.2 Video Frame Extraction *The video frame extracation is done by the scripts
*eus_main.pyandliver_main.pyand their configs are stored in the fileseus_config.yamlandliver_config.yaml3.3 Pre-processing and metadata calculation The preprocessing script takes the extracted frames and saves them after preprocessing. It also creates the metadata files for the preprocessing. Run the script using
pre-process.pyand the project and configs for it should be set inpreprocess_config.yaml.
Important Paths:
EUSML:
- Raw video Directory: /data/eusml/raw_data
- Master Dataset: /data/eusml/master-dataset
- Patient, labels and metadata CSVs: /data/eusml/master-dataset/Data
- Patient Frame CSVs: /data/eusml/master-dataset/Frames
Fatty Liver:
- Raw video Directory: /data/fatty_liver/raw_data
- Master Dataset: /data/fatty_liver/master-dataset
- Patient, labels and metadata CSVs: /data/fatty_liver/master-dataset/Data
- Patient Frame CSVs: /data/fatty_liver/master-dataset/Frames
Support Scripts
For detailed information about support scripts, tools, and applications, see the Support Scripts page.